All-atom simulations unravel the molecular basis of pathogenic mutations and small molecules binding
Fecha: .
Hora: 12:30h.
Lugar: Aula A22.
Ponente: Angelo Spinello (Scienze e Tecnologie Biologiche Chimiche e Farmaceutiche, Università degli Studi di Palermo).
Nowadays, all-atom simulations are widely used to investigate the interactions of small molecules with disease-related biological macromolecules. They are also employed to study their functional dynamics along with the pathological mutations involved in cancer onset. Moreover, they hold great value in guiding and rationalizing experimental campaigns and strikingly improving their chances of success. In this seminar, we will introduce the most important in silico methodologies that have demonstrated an essential role in the field, along with examples taken from the research activity recently performed by our group. These tools may help pave the way toward a calibrated precision medicine approach, leading to drugs that are tailored to the specific patient’s genetic profile.
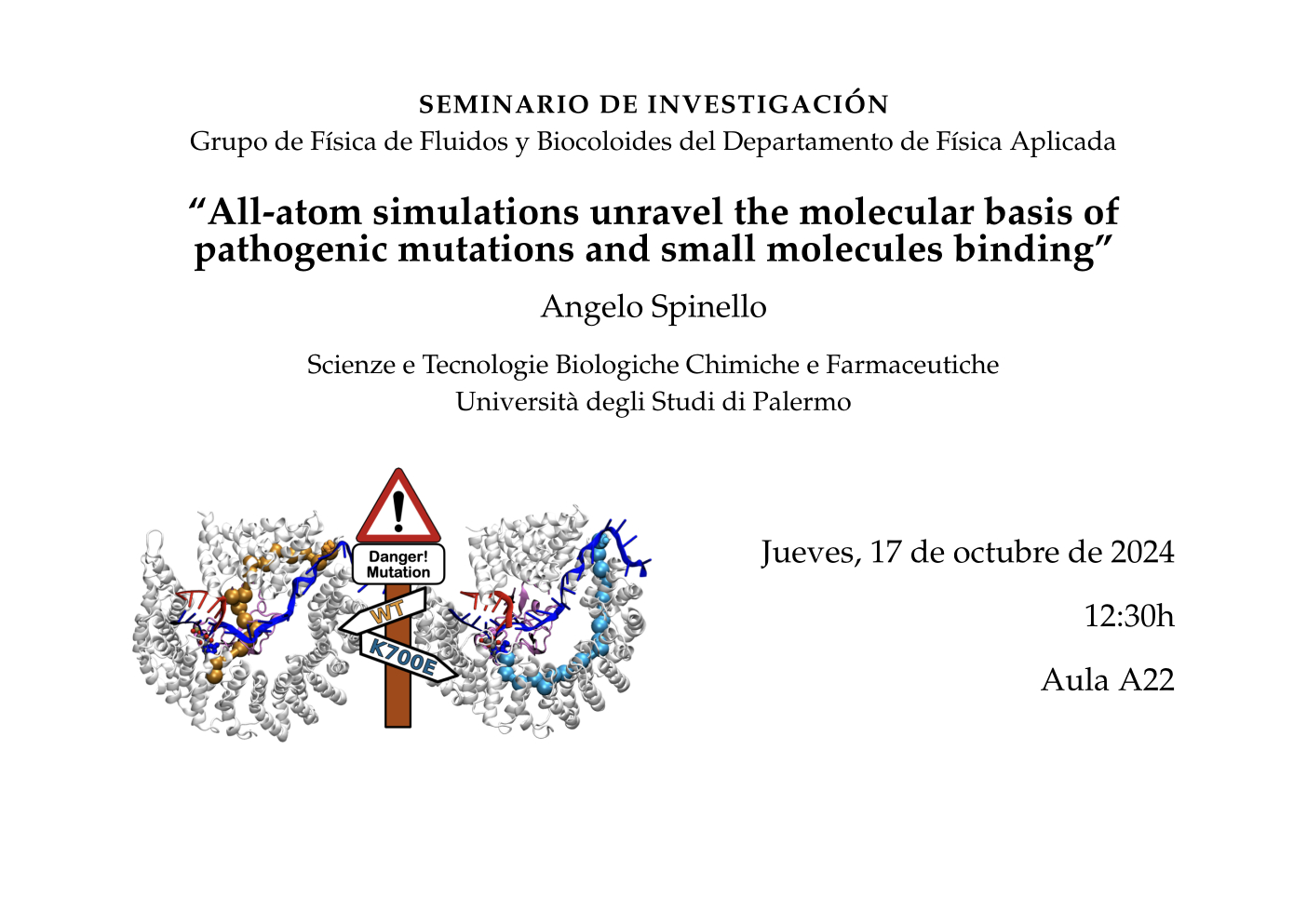
As an example, these will regard the application of molecular dynamics simulations and enhanced sampling methods to rationalize the molecular basis for the preferential lethality exerted by H3B-8800, a small molecule currently in clinical trials, toward mutant cancer cells and the application of dynamical network analysis to unravel the structural basis caused by pathogenic mutations.
El seminario será impartido en inglés.
Contacto: Irene Adroher-Benítez .
Organiza el Grupo de Física de Fluidos y Biocoloides del Departamento de Física Aplicada de la UGR.